Mutations in the genome of a virus can complicate attempts to combat viral diseases, whether by causing a higher number of false negatives in diagnosis methods, such as by PCR, or attenuating or eliminating the effects of anti-viral medication or vaccination by disrupting the pharmacokinetic interactions of viral proteins with administered substances or preventing antibodies from recognizing the target protein. Therefore, it is important to identify the mutatome of SARS-CoV-2, the virus causing the COVID-19 pandemic in-depth and up-to-date, in order to prevent any mutations from stymieing the efforts against COVID-19. To supply researchers in the İzmir Biomedicine and Genome Center and in other research centers with a valuable and easy-to-access resource of the SARS-CoV-2 mutatome and its effects on the viral proteome, we have compared the genomes of 17 SARS-CoV-2 isolates in Turkey that have been sequenced to the reference genome sequence, and analyzed the mutations present in the spike protein region and resulting polypeptide sequence, due to the importance of the protein in recognizing human cell membrane and causing human infections.
Our results are as follows:
* No nonsense mutations are observed, with all isolates capable of producing full-length surface glycoprotein.
* Three distinct mutations are identified. One isolate contains a V772I mutation, with no other mutations. 10 isolates contain a D614G mutation, with no other mutations. One isolate contains both a D614G mutation, and an N679K mutation (Figure 1).
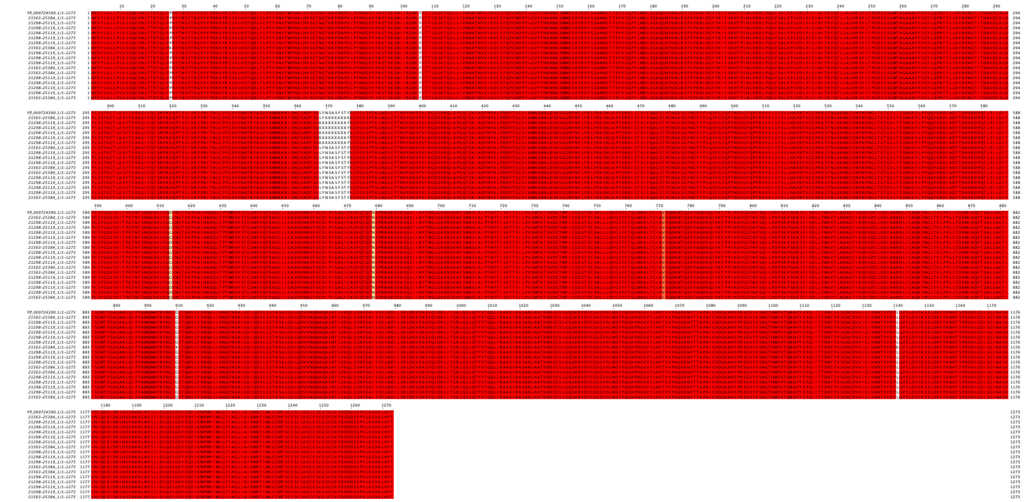
Figure 1. Multiple alignment of spike protein sequences of Turkish isolates. See full alignment in high resolution.
* No indel or inversion mutations are observed.
* All observed mutations are far from spike’s ACE2 binding region. They are also at non-functional sites of the protein, as can be observed from Figure 2.
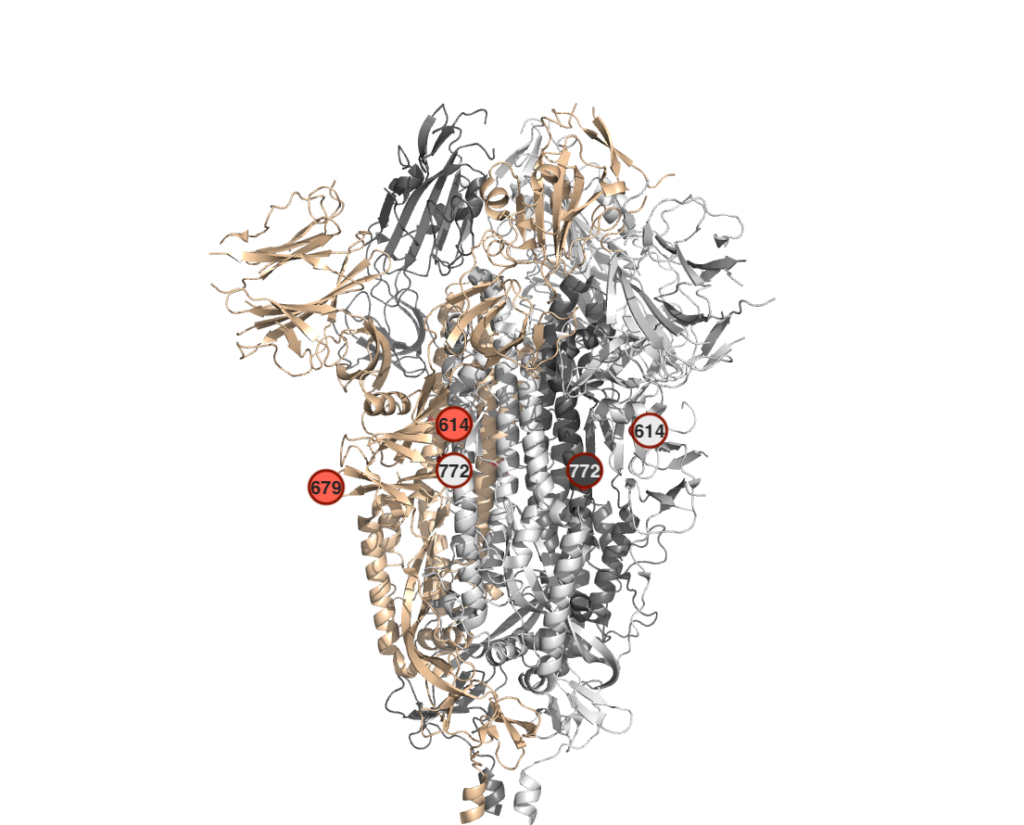
Figure 2. The spike trimer monomers (pdb id: 6vxx) are colored as tint, white and gray. The mutation positions mapped on each monomer are depicted with a circle, including its amino acid position.
* Six isolates have unidentified nucleotides in their sequenced genomes, resulting in unknown amino acid sequences in residues 368-377. The region is identical to the reference genome in all other isolates.
* One isolate has similarly unidentified nucleotides, resulting in unknown amino acid sequences in residues 26, 106, 910, and 1141. These residues are identical to the reference genome in all other isolates.
* Download SARS-CoV-2 spike protein sequences of the isolates: – amino acid sequences – nucleic acid sequences
* Materials and methods can be obtained here
The following researches have contributed to the research summarized above, listed alphabetically in order of surname:
ESKİER, Doğa
KARACA, Ezgi, PhD
KARAKÜLAH, Gökhan, PhD
PAVLOPOULOU, Athanasia, PhD